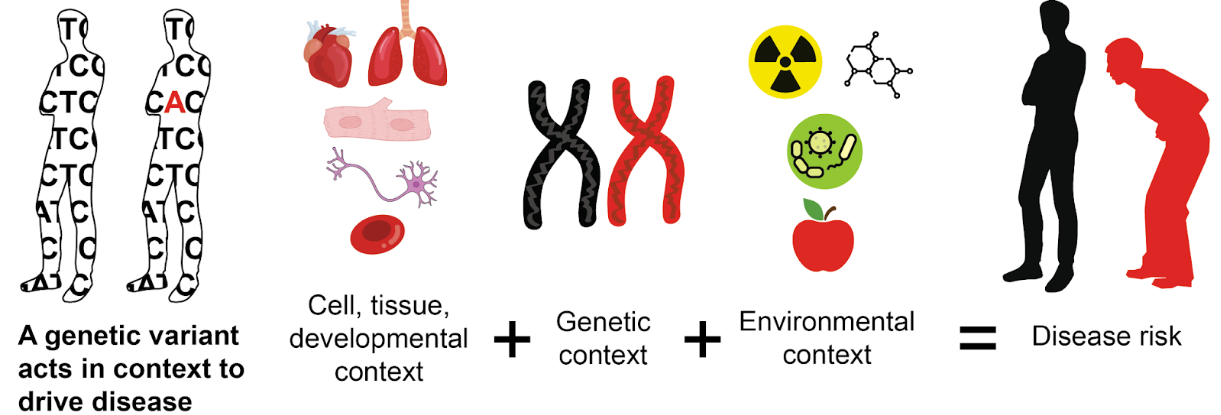
Our experimental and computational approaches will massively increase the return on sequencing an individual’s genome by enabling more accurate variant reinterpretation. We have three guiding aims directing our work.
AIM 1: Systematically assess variant effects in relevant environmental, cell, tissue, and developmental contexts.
We are developing scalable variant effect mapping tools for use in induced pluripotent stem (iPS) cells, organoids and animal models to measure variant effects in specific cell types and multicellular contexts, as well as in varied environments. We are also developing a framework to transfer knowledge of variant effects measured at different scales across environmental contexts.
AIM 2: Systematically assess variant impacts in relevant genetic contexts.
We are developing scalable variant effect mapping tools for use in cell and animal models to measure how variant effects are modified by other variants throughout the genome. Additionally, we are developing a framework to transfer knowledge of variant effects measured at different scales across genetic contexts.
AIM 3:
Exploit context-aware variant effect maps
to infer individualized risk.
We are developing scalable tools combining context-aware variant effect maps with the emerging wealth of human genotype, phenotype, and inferred environmental context to estimate individualized disease risk.